1
NILE: Fast Natural Language Processing for Electronic Health
Records
Sheng Yu
1,2,3,*
, Tianrun Cai
4,5
, Tianxi Cai
6,7,5
1
Center for Statistical Science, Tsinghua University, Beijing, China;
2
Department of Industrial Engineering, Tsinghua University, Beijing, China;
3
Institute for Data Science, Tsinghua University, Beijing, China;
4
Brigham and Women's Hospital, Boston, MA, USA;
5
VA Boston Healthcare System, Boston, MA, USA;
6
Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA,
USA;
7
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA;
* Correspondence to:
Sheng Yu
Weiqinglou Rm 209
Center for Statistical Science
Tsinghua University
Beijing, 100084, China
Email: syu@tsinghua.edu.cn
Tel: +86-10-62783842
Keywords: electronic health records, natural language processing.
2
ABSTRACT
Objective: Narrative text in Electronic health records (EHR) contain rich information for medical
and data science studies. This paper introduces the design and performance of Narrative
Information Linear Extraction (NILE), a natural language processing (NLP) package for EHR
analysis that we share with the medical informatics community.
Methods: NILE uses a modified prefix-tree search algorithm for named entity recognition, which
can detect prefix and suffix sharing. The semantic analyses are implemented as rule-based finite
state machines. Analyses include negation, location, modification, family history, and ignoring.
Result: The processing speed of NILE is hundreds to thousands times faster than existing NLP
software for medical text. The accuracy of presence analysis of NILE is on par with the best
performing models on the 2010 i2b2/VA NLP challenge data.
Conclusion: The speed, accuracy, and being able to operate via API make NILE a valuable addition
to the NLP software for medical informatics and data science.
INTRODUCTION
Electronic health records (EHR) are a rich source of clinical information that can substantially
support biomedical research and healthcare improvement[1,2]. In addition to the structured data
such as lab values and billing codes, EHR consist of clinical narratives recorded by healthcare
providers that typically contain the majority of the encounter details. The unstructured nature of the
narrative data necessitates using natural language processing (NLP) technologies to extract and
standardize the information for analysis. Medical language processing (MLP) systems are specially
designed NLP systems for clinical narratives to handle their distinct syntax and to extract domain-
specific semantics. They are the upstream infrastructure that extract data for researches and
applications ranging from EHR-based phenotyping[3–8] to clinical decision support systems[9,10].
Thus, the efficiency, accuracy, and flexibility of MLP is critically important. NILE is an MLP
system that has been a backbone of our EHR data processing capability and has been used in a
3
variety of research projects.[11–18] We have released NILE to the research community, and in this
paper we introduce its design and performance.
A complete MLP workflow typically involves two major steps: named entity recognition and
semantic analysis. Named entity recognition (NER) is the process of identifying mentions of
concepts in the text, such as diseases, symptoms, and medications. One commonly used approach
for NER is “chunk-and-match”. Chunking is the process to identify phrases of interest (usually
noun phrases). Many MLP programs, including HITEx[19], cTAKES[20], MetaMap[21], and
KnowledgeMap[22], do part-of-speech (POS) tagging before chunking, and use the POS to
determine the phrase boundaries. CAPIS[23] uses a simple heuristic to find phrase boundaries.
LEXIMER[24] does phrase separation via punctuation and entropy reduction. An extension of
CLARIT[25] also employs corpus statistics for the identification of phrases. After chunking,
various term identification methods are used to match the phrases against their dictionaries to see
if they are recognizable. Another convenient tool for simple term identification is regular
expressions. NegEx and ConText[26,27] use regular expressions to identify cues for semantic
analysis. HITEx also has a pipeline that uses regular expressions to identify medications from
discharge notes. However, simple implementations of regular expressions search for dictionary
terms one by one. As a result, the processing speed grows linearly with the size of the dictionary.
Therefore, regular expressions are not suitable for EHR processing using a large dictionary like the
Unified Medical Language System (UMLS)[28]. An efficient alternative to regular expression
based methods is tree-based search algorithms which tend to be extremely fast in speed. The Aho-
Corasick algorithm[29] used in MedTagger[30] is an example of such algorithms. NILE also uses
tree-based search and is capable of handling prefix and suffix sharing that are common in EHR.
Semantic analysis usually uses the output of the NER step as input and annotates them by the
meaning in the text. Common analyses for named entities (symptoms, medications, etc.) include
whether they are present/used, conditional, or whether they are hypothetical or part of either the
4
past medical history or family history. Common analyses for attributes of a condition or symptom
include determining its location, severity, and chronicity. There are other specialized analyses, such
as determining the smoking state of the patient[31,32], value extraction from semi-structured
EHR[33,34], extraction of drug and food allergies[35] and medication information[36]. The
analysis can be based on rules or be data-driven using machine learning. A number of famous
systems, including MedLEE[37], NegEx[26], and ConText[27], use rules based on patterns of cue
words. NILE is also rule-based for robustness and efficiency. When annotated sentences are
available, machine learning is another good option. The best contenders in the 2010 i2b2/VA
challenge unanimously used machine learning for semantic analysis, employing support vector
machines as well as other techniques, such as conditional random fields and logistic
regressions.[38–45]
There are several reasons for why NILE can be a valuable addition to existing publicly available
software such as cTAKES and MetaMap. First, instead of being a software operated via a graphical
user interface, NILE takes the form of a Java package. EHR are stored in databases with different
schemes, and notes are usually written in diverse formats and templates. So, note parsing usually
require customization. A package therefore can be more appealing to data scientists who want to
easily write a piece of code to command how to query the EHR database, how to segment the notes,
when and where the NLP should kick in, and how the results should be formatted. Second, the
UIMA framework[46] adopted by cTAKES and other software is very hard for non-NLP
professionals and requires significant time to learn. In contrast, the concepts and APIs used in NILE
are easily understood by people who know object-oriented programming. Finally, NILE is
extremely fast. Our tests show that NILE is by several orders of magnitude faster than existing
software, which is critical to analyzing large EHR databases. At Partners HealthCare and Veterans
Affairs, we have used NILE to analyze billions of notes. NILE is currently provided in two forms
– a Java package operated via APIs, and a Windows executable incorporating additional features
such as multi-threading and database access that can be operated by people without programming

5
background. Download is available at
http://celehs.hms.harvard.edu/software/NILE/NILE_main.html.
DESIGNS AND FUNCTIONS
NILE stands for the Narrative Information Linear Extraction system. The system is called “linear”
because NILE’s NER and semantic analyzers work by sweeping through the sentence from left to
right, allowing NILE to be extremely fast. Medical note processing with NILE typically involves
the following steps: 1. Initiate NILE and load the dictionary; 2. Read a note and preprocess it to
identify paragraphs that need NLP analysis; 3. Give a paragraph to NILE, which will segment the
paragraph to sentences and tokenize them; 4. Apply NER to each sentence; 5. Apply semantic
analyses; 6. Write the result to the output device. Step 2 requires the user to understand the target
notes and customize for them. Steps 3-5 are packed in a single command for simplicity. Step 6
allows the user to decide what parts of the result are needed and to customize the output format.
The rest of this section introduces concepts and designs of NILE’s NER and semantic analysis
components.
Named Entity Recognition
The dictionary entries in NILE are called Phrases, which are objects that contain not only the text,
but also their semantic roles and codes. There are broadly two kinds of Phrases: 1) grammatical
words and meaning cues that we have collected and included as part of the NILE package, and 2)
medical terms that need to be provided by the user. For example, the user can load the dictionary
with terms extracted from the UMLS. Each Phrase has exactly one semantic role that tells the
semantic analyzers how to use the phrase. Currently there are 29 predefined semantic roles, 26 of
which are for grammatical words and meaning cues, and 3 (observation, modifier, and location) are
6
for medical terms. When modification information is not needed, which is a common case in note
parsing, the user only needs to provide observation terms, which are typically nouns or noun phrases
include diseases, symptoms, findings, medications, procedures, etc. A code is used as a concept ID
for the Phrase. Typically, if one uses terms extracted from the UMLS, then the Concept Unique
Identifier (CUI) will naturally be the code. A Phrase is allowed to have multiple codes. This design
for one reason is to reflect the fact that many terms have coding ambiguities. For instance, UMLS
concepts C0010054, C0010068, and C1956346 are very close and have common terms “coronary
artery disease” and “coronary heart disease”. Therefore, when a term with ambiguous codes is
identified, the user may wish to output all the codes. Another use case of the multiple codes is to
reflect the concept hierarchy. For example, we can find from the UMLS concept relation table that
Tylenol (C0699142) is a tradename of Paracetamol (C0000970) and that Paracetamol is a kind of
Analgesics (C0002771). Thus, we can let the entry “Tylenol” have all three codes, and when
“Tylenol” is identified in a note, all three codes are written to the output. This facilitates concept
aggregation and allows one to do flexible feature selection, as seen in automated feature selection
for high-throughput phenotyping.[12,13]
The dictionary of NILE uses a prefix tree data structure. Figure 1 illustrates a prefix tree with five
terms: “heart”, “heart failure”, “heart attack”, “coronary angiogram”, and “coronary artery”. If a
term ends at a node, then the node contains the corresponding Phrase object; otherwise that field is
null. Each node uses a hash table to connect to its children via the next token (word) in the term.
The hash tables make the search speed insensitive to the number of entries in the dictionary. The
matching algorithm reads a sentence as a series of tokens, and matches the longest phrase from left
to right. For instance, when NILE sees “patient had a heart attack in 2006…” it identifies “heart
attack” rather than “heart”, and goes on to find the next identifiable phrase starting from “in”. We
extended the basic prefix tree search algorithm to recognize prefix and suffix sharing.
Supplementary Material Algorithm 1 is the pseudo code that returns the longest phrase starting at
the i-th token if the first argument node is the root. The biggest if-block extends the basic algorithm

7
to recognize suffix sharing, so that “mediastinal, hilar, or axillary lymphadenopathy” can be
recognized as “mediastinal lymphadenopathy, hilar lymphadenopathy, or axillary
lymphadenopathy”. Prefix sharing is handled in the following way: if the previous identified term
is at least two words long, keep the first word as prefix; if matching at the next position fails, try
again with match(root.child(prefix), tokens, i). The prefix is reset to empty if
the match fails again or returns the prefix itself. With both prefix and suffix sharing recognition, we
can now interpret “right upper, middle, and lower lobes” as “right upper lobe, right middle lobe,
and right lower lobe”.
NILE’s NER is case sensitive. In general situations, we encourage the user to convert both the
dictionary terms and the input sentences to lowercase. The cases in clinical notes can be very
unpredictable – some notes or paragraphs are written entirely in uppercase. So, while lowercasing
everything can bring ambiguity to some acronyms, we find it a good trade-off in general. Another
thing to note is lemmatization, which is a common step in NER, typically achieved via the LVG
lexical tools.[47] In the design of NILE, we have avoided lemmatization to keep the matching
algorithm pure and minimal; and to match plural forms, we expect the user to populate the
dictionary with plural forms using the LVG.
Figure 1: Dictionary data structure
8
Semantic Analysis
The output of the NER step is a list of Semantic Objects, which are essentially Phrases packed with
additional offset information. Words and phrases not in the dictionary are not kept in the NER
output. The semantic analysis step involves a sequence of analyzers with a common API that uses
List<SemanticObject> as both input and output. Each analyzer may modify attributes of the
Semantic Objects (e.g., presence and family history), attach Semantic Objects to other ones
(modification), or remove Semantic Objects from the list. The analyzers are implemented as finite
state machines that look at the Semantic Objects in the list sequentially, modify their states
depending on the current states and what objects they see, and decide their actions on each Semantic
Object. Each analyzer is designed to be simple, so an analysis (e.g., presence analysis and location
analysis) may comprise multiple analyzers. Implemented analyses include presence, location,
modification (other than location), family history, and an ignoring analyzer that will remove an
identified observation object from the output if it does not express presence or absence. For example,
“patient came to rule out rheumatoid arthritis” and “Breast Cancer Center” do not express presence
or absence of “rheumatoid arthritis” or “breast cancer”, so these observation objects will be
removed, and the user will not see them in the output. In the following, we introduce presence and
location analysis in more detail.
Presence Analysis
The presence analysis (also known as negation analysis) analyzes whether a problem is present, a
drug is prescribed/used, a test is taken, etc., and assigns values YES, NO, or MAYBE. The analysis
is performed in two steps (two analyzers), starting with preprocessing and followed by presence
tagging. The preprocessing checks if consecutive meaning cues or grammatical words can form a
new cue. For example, originally in the dictionary, “found” is a past participle for confirmation.
When the analyzer detects a sequence in the left column of Table 1, it replaces the Semantic Objects

9
with a new object with the role in the right column. The analyzer only looks at the semantic roles,
so that the rules can generalize. For example, if “found” is replaced by “identified”, or if “isn’t” is
replaced by “weren’t”, they would appear the same to the analyzer.
Table 1: Combined effects of semantic roles.
Combination
Semantic Role
found (used alone)
Confirmation
not + found
backward negation
have + found
Confirmation
haven' t + found
Negation
have + been + found
backward confirmation
haven't + been + found
backward negation
is + found
backward confirmation
isn't + found
backward negation
After the preprocessing, the presence tagger iterates through the Semantic Objects. When the tagger
sees an observation object, it applies the current presence state to it and puts it in the cache.
Depending on the semantic role or the role transition of the observed objects, the tagger may change
the presence state, change the attributes of the observation objects in the cache (when seeing
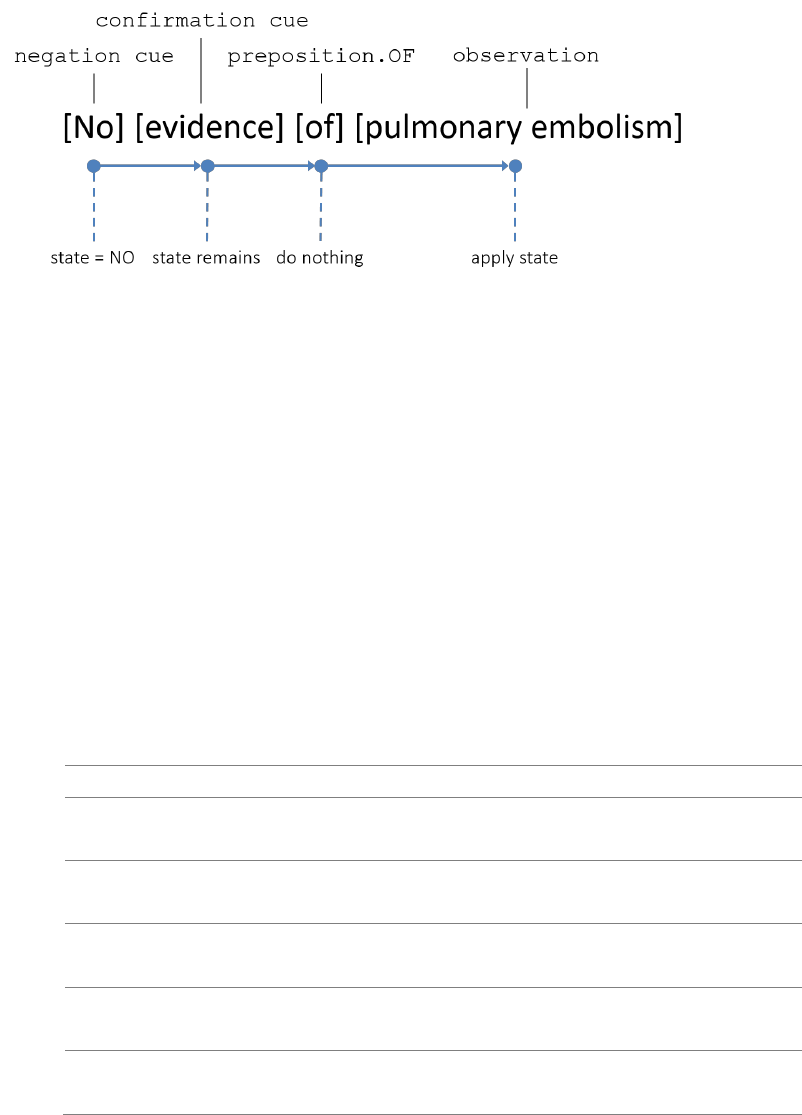
backward cues, e.g., “was not found”), or empty the cache. Figure 2 provides an illustration. The
analyzer initiates with presence state YES and starts from the first semantic object. Since “no” is a
negation cue, the presence state changes to NO. The analyzer then proceeds to “evidence”, which
is a confirmation cue. In the current implementation of the analyzer, negation always suppresses
confirmation, so the state remains as NO. The analyzer does nothing when it sees the preposition
“of” and goes on. Finally, when the analyzer sees the observation object “pulmonary embolism”, it
applies the current state NO to the object’s presence attribute.
10
Figure 2: Illustration of the presence analysis.
With enough granularity of semantic roles and rules, the presence analyzers can correctly classify
the presence most of the times, including subtle differences as shown in Table 2, which are usually
too hard for existing software. We have also added support for presence analysis from other
analyzers. For example, words like “change”, “increasing”, and “worsening” are given a semantic
role that represents change, and an analyzer that precedes presence analysis detects them and their
targets, so that later in “no change in pleural effusion”, the negation will only apply to “change”
but not to “pleural effusion”.
Table 2: Handling subtle differences in presence analysis.
Example Sentence
Return
No filling defects are seen to suggest pulmonary
embolism.
filling defects: NO
pulmonary embolism: NO
No filling defects are seen, suggesting
pulmonary embolism.
filling defects: NO
pulmonary embolism: YES
No filling defects are seen and it suggests
pulmonary embolism.
filling defects: NO
pulmonary embolism: YES
No filling defects that suggest pulmonary
embolism are seen.
filling defects: NO
pulmonary embolism: NO
No filling defects are seen, which suggests
pulmonary embolism.
filling defects: NO
pulmonary embolism: YES
Location Analysis
Identification of problem locations is another useful analysis in MLP. NILE can handle
sophisticated sentences and do nesting analysis for anatomical locations, which is an important
11
feature that is missing in other existing MLP programs.
Descriptions of anatomical locations usually involve nested modifications that can be expressed in
very diverse ways. The simplest patterns of nested modifications are “location A location B” (e.g.,
“left upper lobe arteries”) and “location A of/in/within/... location B” (e.g., “arteries of the left upper
lobe”). The analyzer iterates through the sequence of Semantic Objects, and treats the former
pattern as “location B (location A)” and the latter as “location A (location
B)”, where objects in the parentheses modify the object preceding the parentheses. In the
implementation, the object inside of the parentheses is removed from the Semantic Object list and
its reference is added to the modifier list of the object preceding the parentheses. These patterns
can be chained. For instance, “location A location B location C” is interpreted as “location C
(location B (location A))”. The same patterns are used to assign location objects to the
observation objects that they modify, except that observation objects cannot be modifiers. For
example, “location A fact B” means “fact B (location A)”, and “fact A in location B
location C” means “fact A (location C (location B))”.
The above patterns are simple because they involve a single nesting chain. The analysis becomes
difficult when the modification structure has branches (such as two modifiers joined by “and”). The
following is an example from a computed tomography pulmonary angiography (CTPA) report:
“There are segmental and subsegmental filling defects in the right upper lobe, superior segment of
the right lower lobe, and subsegmental filling defect in the anterolateral segment of the left lower
lobe pulmonary arteries.”
This sentence includes multiple observation objects, each having multiple location modifiers,
whose modification structures contain both nesting and branches. The location analyzer checks the
semantic roles of the current and the previous object as well as the nesting state (forward/backward)

12
to decide whether to nest or to branch. For the above sentence, using a custom CTPA dictionary[11],
NILE returned:
filling defects: YES (right upper lobe; superior segment (right
lower lobe); segmental; subsegmental),
filling defect: YES (segment (pulmonary arteries (left lower lobe));
subsegmental),
which is the same as the intended meaning, except that “anterolateral segment” was not in the
dictionary.
EVALUATION
We provide speed and accuracy evaluations for NILE. The computer for benchmarking the speed
has an AMD Ryzen 7 2700X CPU and 32 GB RAM, running Java 1.8.0_191. The data set used for
testing the presence analysis accuracy is the 2010 NLP i2b2/VA challenge data, available at
https://i2b2.org/NLP/DataSets.
Speed
We compare NILE with cTAKES 4.0.0 and MedTagger 1.0.1 to process 10,330 CTPA reports.
cTAKES is a popular open source MLP system that uses the “chunk-and-match” approach for NER
and uses machine learning based modules for semantic analysis. MedTagger mainly focuses on
NER and uses a prefix-tree search algorithm, so it is expected to be faster than cTAKES, but it does
not have the ability to detect prefix and suffix sharing like NILE. Both cTAKES and MedTagger
were run with their default dictionaries and recommended memory allocation (3 GB for cTAKES,
2 GB for MedTagger). We tested NILE (single-threaded) with two sets of dictionaries: a small
dictionary with about 1000 terms that we curated for a pulmonary embolism study[11], and a large

13
dictionary with an additional 0.9 million terms arbitrarily drawn from the UMLS. Table 3 shows
the average processing time for the 10,330 CTPA reports, where the time for cTAKES and
MedTagger are the average of two runs, and the time for NILE for both situations are the average
of 10 runs.
Table 3: Average processing time for 10,330 CTPA reports.
Software/setting
Processing time
cTAKES 4.0.0
4305.55 sec
MedTagger 1.0.1
816.17 sec
NILE (small dictionary)
1.84 sec
NILE (large dictionary)
2.18 sec
Accuracy of Presence Analysis in Discharge Summaries
The 2010 i2b2/VA NLP challenge dataset contains discharge summaries annotated with the problem
terms of interest and a corresponding label for their presence with six possible values: “present”,
“absent”, “possible”, “hypothetical”, “conditional”, and “associated_with_someone_else”. To
accommodate for these annotations, we customized NILE and combined several analyzers to jointly
classify the presence, with the exception of “conditional”, as we did not have an analyzer for it. The
dataset contains 170 reports for training and 256 reports for testing, which are fewer than the
reported size in the 2010 challenge (394 and 477 reports for training and testing, respectively).[45]
NILE’s analyzers are rule-based and do not need training. Instead, we used the training data to
understand the annotator’s logic and reassigned the semantic roles of some of the cue words
accordingly. This customized version is released together with the standard version of NILE. We
encourage using the standard version because it suits more general scenarios (see Supplementary
Material for details).

14
Table 4 shows the F scores of NILE for classifying the presence state on the test data, with
comparison to the reported scores of deBruijn[38] (system 2.3) and MITRE[39], which ranked 1
st
and 2
nd
, respectively, in the 2010 i2b2/VA challenge. The confusion matrices are given in the
Supplementary Material.
Table 4: F scores for classifying the presence state on the 2010 i2b2/VA challenge data.
NILE
deBruijn
MITRE
absent
0.934
0.929
0.94
someone_else
0.954
0.899
0.87
conditional
--
0.355
0.42
hypothetical
0.860
0.883
0.89
possible
0.627
0.627
0.63
present
0.954
0.952
0.96
DISCUSSION
NILE is a time tested software initially developed in 2012 and has been used in many projects over
the years[11–18]. The most important merit of NILE is that it is significantly faster than other
publicly available platforms while maintaining a comparable accuracy. As Table 3 shows, NILE is
2000 times faster than cTAKES and 400 times faster than MedTagger, and the dictionary size
minimally affect the processing speed. As the amount of EHR data available for analysis keeps
growing, a large processing bandwidth has become a critical quality to have for medical data
science. The speed of NILE has given us the capability to process very large EHR data sets, such
as performing phenome-wide association studies (PheWAS) using million-patient scale EHR data
from the VA MVP[48]. In practice, we find that the majority of processing time is spent on the I/O
(reading database and writing the result) rather than on the NLP, so we did not incorporate multi-
threading for NILE in the core package. However, a Windows executable version with multi-
15
threading capability is available for download.
Choosing rule-based semantic analysis over machine learning is another design decision. While
NLP improves rapidly with the current research of deep learning, the availability of large-scale
annotated data is a major obstacle to widely using machine learning. In comparison, the
development of rule-based semantic analyzers is not limited by the availability of annotated data,
and their performances are generally acceptable. Table 4 shows that the accuracy of NILE’s
presence analysis is on par with the top performing machine learning models from the i2b2/VA
challenge. As detailed in the Supplementary Material, NILE’s presence analyzer have different
annotation labels and logic from that used in the challenge. Being able to customize NILE for this
benchmark also reflects NILE’s flexibility from being a package operated via API, which can be
appealing to data scientists in medical informatics.
From the experience of using NILE, we have found a few limitations of it that are worth noting.
One limitation is that the values of the presence state (YES, NO, or MAYBE) do not fully cover
the possibilities, especially the time dimension. For example, in “the patient has ceased smoking”
and “the symptom has disappeared”, simply assigning YES or NO cannot fully record the
information from the sentences. Another limitation is term sense disambiguation. Acronyms often
have multiple possible meanings. For example, “CHD” could represent congenital heart disease,
coronary heart disease, or child/children; “DR” could be doctor, drive, delayed release, or diabetic
retinopathy; “RA” could be rheumatoid arthritis or room air (for testing oxygen saturation). NILE
does not do disambiguation analysis. However, the problem can be addressed outside of NILE, e.g.,
using distributional semantics[49,50].
CONCLUSION
In this paper, we introduced the design and performance of NILE, an NLP package for EHR analysis
16
that we share with the medical informatics community. NILE is hundreds to thousands times faster
than existing software, with accuracy on par with the best performing models in the 2010 i2b2/VA
challenge. These qualities along with being able to program via API make it an appealing addition
to the existing NLP software for data scientists in this field.
Funding: None.
Competing Interests: None.
Contributorship: Sheng Yu developed the original NILE as a Java package. Tianrun Cai developed
the Windows executable version with additional features including multi-threading and database
access. Tianxi Cai provided guidance on the design and testing of the NILE package. All authors
contributed to evaluating the performance of NILE in analyzing EHR data and drafting and
critically revising this manuscript.
REFERENCES
1 Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev
Genet 2011;12:417–28. doi:10.1038/nrg2999
2 Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual
documents in the electronic health record: a review of recent research. Yearb Med Inform
2008;:128–44.
3 Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohnʼs disease and
ulcerative colitis in electronic medical records using natural language processing: a novel
informatics approach. Inflamm Bowel Dis 2013;19:1411–20.
doi:10.1097/MIB.0b013e31828133fd
4 Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid
arthritis. Arthritis Care Res 2010;62:1120–7. doi:10.1002/acr.20184
5 Kumar V, Liao K, Cheng S-C, et al. Natural language processing improves phenotypic
accuracy in an electronic medical record cohort of type 2 diabetes and cardiovascular disease.
J Am Coll Cardiol 2014;63:A1359. doi:10.1016/S0735-1097(14)61359-0
17
6 Love TJ, Cai T, Karlson EW. Validation of psoriatic arthritis diagnoses in electronic medical
records using natural language processing. Semin Arthritis Rheum 2011;40:413–20.
doi:10.1016/j.semarthrit.2010.05.002
7 Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid
arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162–9.
doi:10.1136/amiajnl-2011-000583
8 Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using
electronic health records. PLoS ONE 2013;8:e78927. doi:10.1371/journal.pone.0078927
9 Lindberg DAB, Rowland LR, Buch CRJr, et al. CONSIDER: A computer program for medical
instruction. In: Proceedings of 9th IBM Medical Symposium. 1968. 54.
10 Blois MS, Tuttle MS, Sherertz DD. RECONSIDER: a program for generating differential
diagnoses. In: Proceedings of the Fifth Annual Symposium on Computer Applications in Health
Care. 1981. 263–8.
11 Yu S, Kumamaru KK, George E, et al. Classification of CT pulmonary angiography reports by
presence, chronicity, and location of pulmonary embolism with natural language processing. J
Biomed Inform 2014;52:386–93. doi:10.1016/j.jbi.2014.08.001
12 Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated
feature extraction and selection from knowledge sources. J Am Med Inform Assoc
2015;22:993–1000. doi:10.1093/jamia/ocv034
13 Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput
phenotyping. J Am Med Inform Assoc 2017;24:e143–9. doi:10.1093/jamia/ocw135
14 Geva A, Gronsbell JL, Cai T, et al. A Computable Phenotype Improves Cohort Ascertainment
in a Pediatric Pulmonary Hypertension Registry. J Pediatr 2017;188:224-231.e5.
doi:10.1016/j.jpeds.2017.05.037
15 Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform
Assoc 2018;25:54–60. doi:10.1093/jamia/ocx111
16 Beam AL, Kompa B, Fried I, et al. Clinical Concept Embeddings Learned from Massive
Sources of Multimodal Medical Data. ArXiv180401486 Cs Stat Published Online First: 4 April
2018.http://arxiv.org/abs/1804.01486 (accessed 5 Oct 2018).
17 Cai T, Lin T-C, Bond A, et al. The Association Between Arthralgia and Vedolizumab Using
Natural Language Processing. Inflamm Bowel Dis 2018;24:2242–6. doi:10.1093/ibd/izy127
18 Gronsbell J, Minnier J, Yu S, et al. Automated feature selection of predictors in electronic
18
medical records data. Biometrics 2019;75:268–77. doi:10.1111/biom.12987
19 HITEx Manual. https://www.i2b2.org/software/projects/hitex/hitex_manual.html (accessed 14
Jan 2014).
20 Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge
Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med
Inform Assoc 2010;17:507–13. doi:10.1136/jamia.2009.001560
21 Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap
program. Proc AMIA Symp 2001;:17.
22 Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content
using KnowledgeMap. J Am Med Inform Assoc 2003;10:351–62. doi:10.1197/jamia.M1176
23 Lin R, Lenert L, Middleton B, et al. A free-text processing system to capture physical findings:
Canonical Phrase Identification System (CAPIS). Proc Annu Symp Comput Appl Med Care
1991;:843.
24 Dreyer KJ, Kalra MK, Maher MM, et al. Application of Recently Developed Computer
Algorithm for Automatic Classification of Unstructured Radiology Reports: Validation Study.
Radiology 2005;234:323–9. doi:10.1148/radiol.2341040049
25 Evans DA, Zhai C. Noun-phrase Analysis in Unrestricted Text for Information Retrieval. In:
Proceedings of the 34th Annual Meeting on Association for Computational Linguistics.
Stroudsburg, PA, USA: : Association for Computational Linguistics 1996. 17–24.
doi:10.3115/981863.981866
26 Chapman WW, Bridewell W, Hanbury P, et al. A Simple Algorithm for Identifying Negated
Findings and Diseases in Discharge Summaries. J Biomed Inform 2001;34:301–10.
doi:10.1006/jbin.2001.1029
27 Harkema H, Dowling JN, Thornblade T, et al. ConText: An algorithm for determining negation,
experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51.
doi:10.1016/j.jbi.2009.05.002
28 Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between
users and the information they need. Bull Med Libr Assoc 1993;81:170.
29 Aho AV, Corasick MJ. Efficient String Matching: An Aid to Bibliographic Search. Commun
ACM 1975;18:333–340. doi:10.1145/360825.360855
30 Liu H, Wagholikar KB, Jonnalagadda S, et al. Integrated cTAKES for Concept Mention
Detection and Normalization. In: CLEF (Working Notes). 2013.
19
31 Uzuner Ö, Goldstein I, Luo Y, et al. Identifying Patient Smoking Status from Medical
Discharge Records. J Am Med Inform Assoc 2008;15:14–24. doi:10.1197/jamia.M2408
32 Clark C, Good K, Jezierny L, et al. Identifying Smokers with a Medical Extraction System. J
Am Med Inform Assoc 2008;15:36–9. doi:10.1197/jamia.M2442
33 Turchin A. Identification of clinical characteristics of large patient cohorts through analysis
of free text physician notes. 2005.http://dspace.mit.edu/handle/1721.1/33085 (accessed 22 Jul
2014).
34 Kristianson KJ, Ljunggren H, Gustafsson LL. Data extraction from a semi-structured electronic
medical record system for outpatients: A model to facilitate the access and use of data for
quality control and research. Health Informatics J 2009;15:305–19.
doi:10.1177/1460458209345889
35 Epstein RH, Jacques PS, Stockin M, et al. Automated identification of drug and food allergies
entered using non-standard terminology. J Am Med Inform Assoc 2013;20:962–8.
doi:10.1136/amiajnl-2013-001756
36 Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for
clinical narratives. J Am Med Inform Assoc 2010;17:19–24. doi:10.1197/jamia.M3378
37 Friedman C, Alderson PO, Austin JHM, et al. A General Natural-language Text Processor for
Clinical Radiology. J Am Med Inform Assoc 1994;1:161–74.
doi:10.1136/jamia.1994.95236146
38 deBruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of
clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc
2011;18:557–62. doi:10.1136/amiajnl-2011-000150
39 Clark C, Aberdeen J, Coarr M, et al. MITRE system for clinical assertion status classification.
J Am Med Inform Assoc 2011;18:563–7. doi:10.1136/amiajnl-2011-000164
40 Minard A-L, Ligozat A-L, Abacha AB, et al. Hybrid methods for improving information access
in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc
2011;18:588–93. doi:10.1136/amiajnl-2011-000154
41 Xu Y, Hong K, Tsujii J, et al. Feature engineering combined with machine learning and rule-
based methods for structured information extraction from narrative clinical discharge
summaries. J Am Med Inform Assoc 2012;19:824–32. doi:10.1136/amiajnl-2011-000776
42 Roberts K, Rink B, Harabagiu SM, et al. A Machine Learning Approach for Identifying
Anatomical Locations of Actionable Findings in Radiology Reports. AMIA Annu Symp Proc
2012;2012:779.
20
43 Dligach D, Bethard S, Becker L, et al. Discovering body site and severity modifiers in clinical
texts. J Am Med Inform Assoc 2013;:amiajnl-2013-001766. doi:10.1136/amiajnl-2013-001766
44 Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts
in clinical texts. J Am Med Inform Assoc 2011;18:594–600. doi:10.1136/amiajnl-2011-000153
45 Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and
relations in clinical text. J Am Med Inform Assoc 2011;:amiajnl-2011-000203.
doi:10.1136/amiajnl-2011-000203
46 Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing
in the corporate research environment. Nat Lang Eng 2004;10:327–48.
doi:10.1017/S1351324904003523
47 McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical
terminologies. Proc Annu Symp Comput Appl Med Care 1994;:235–9.
48 Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study
genetic influences on health and disease. J Clin Epidemiol 2016;70:214–23.
doi:10.1016/j.jclinepi.2015.09.016
49 Mikolov T, Sutskever I, Chen K, et al. Distributed Representations of Words and Phrases and
their Compositionality. In: Burges CJC, Bottou L, Welling M, et al., eds. Advances in Neural
Information Processing Systems 26. Curran Associates, Inc. 2013. 3111–3119.
50 Pennington J, Socher R, Manning CD. Glove: Global vectors for word representation. In: In
EMNLP. 2014.

21
Supplementary Material
NILE: Fast Natural Language Processing for Electronic Health
Records
Sheng Yu
1,2,3,*
, Tianrun Cai
4,5
, Tianxi Cai
6,7,5
1
Center for Statistical Science, Tsinghua University, Beijing, China;
2
Department of Industrial Engineering, Tsinghua University, Beijing, China;
3
Institute for Data Science, Tsinghua University, Beijing, China;
4
Brigham and Women's Hospital, Boston, MA, USA;
5
VA Boston Healthcare System, Boston, MA, USA;
6
Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA,
USA;
7
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA;
* Correspondence to:
Sheng Yu
Weiqinglou Rm 209
Center for Statistical Science
Tsinghua University
Beijing, 100084, China
Email: syu@tsinghua.edu.cn
Tel: +86-10-62783842
Keywords: electronic health records, natural language processing.
Prefix tree search with suffix-sharing recognition
function match(node, tokens, i) {
if (node == null) return null
if (i >= tokens.length) return null

22
p = match(node.child(tokens[i]), tokens, i + 1)
if (p != null) return p
if (tokens[i] == "and" OR "or" OR ",")
k = LIMIT // a constant integer, e.g., 3 or 4
for (j = i + 2; j < tokens.length AND k > 0; j++, k--)
if (tokens[j] == "and" OR "or" OR ",")
k = LIMIT
continue
if (node.child(tokens[j]) != null)
p = match(node, tokens, j)
break
if (p != null) return p
return node.phrase
}
Algorithm 1: Prefix tree search with suffix-sharing recognition
Customization and Results for the 2010 i2b2/VA
challenge data
The 2010 i2b2 NLP data set contains clinical narrative texts that have been chunked to sentences,
i.e., ideally each line contains one complete sentence. Each sentence has also been tokenized with
tokens separated by space. The reference data specifies the problem terms of interest and provides
a value for their presence, which could be “present”, “absent”, “possible”, “hypothetical”,
“conditional”, or “associated_with_someone_else”. The “present”, “absent”, and “possible”
correspond to the YES, NO, and MAYBE values in our presence analyzer. The value “hypothetical”
is given in cases such as “call if you experience shortness of breath”. It is similar to our analyzer
for ignoring, thus we modified it to detect the hypothetical problems. The value “conditional” is
given when the sentence states that a problem can be triggered by certain conditions. We do not
have similar analysis, thus we removed them from the test. The value
“associated_with_someone_else” means family history most of the times, and we used our family
history analyzer for it.
One issue of the reference is that it also considers the meaning of the specified terms. For instance,
a term could be “afebrile”, meaning “no fever”, and the reference value for presence would be
“absent”. The way NILE works is to assign YES to the presence state to “afebrile” rather than NO
to “fever”. Thus, to accommodate this problem, we added a postprocessor and forced the presence
of a term to be “absent” if it was known to express negation by itself. Another ad hoc customization

23
was the detection of the family history section, and all problem terms in the section were labeled
as “associated_with_someone_else”.
The data set includes training data and testing data. Our analyzers are rule-based and do not need
training. Instead, we read and test ran with the training data to understand the annotator’s logic and
modified our cue words accordingly. For instance, normally we use “evaluate” and “rule out” as
cues to ignore a part of a sentence, but in this test we made them speculation cues so that their
objects would be labeled as “possible”. The terms that self-expressed negation were also found with
the test run.
This customized version of NILE is available upon request.
Prediction
Reference
absent
someone_else
hypothetical
possible
present
sensitivity
absent
1527
1
3
4
62
0.956
someone_else
1
82
1
0
5
0.921
hypothetical
3
0
344
1
34
0.901
possible
15
0
4
195
95
0.631
present
77
7
29
77
4432
0.959
precision
0.941
0.911
0.903
0.704
0.958
Table 5: Confusion matrix for the training data set
Prediction
Reference
absent
someone_else
hypothetical
possible
present
sensitivity
absent
2434
3
5
8
146
0.938
someone_else
1
125
0
0
5
0.954
hypothetical
6
0
400
5
34
0.899
possible
21
0
16
366
249
0.561
present
154
3
64
136
8259
0.959
precision
0.930
0.954
0.825
0.711
0.950
Table 6: Confusion matrix for the testing data set
